
Alignments are obtained by combining, weighting and screening the results of several multiple alignment programs. 2010. Nature Protocols 5: 725-738)ĮSyPred3D - this automated homology modeling program derives benefit from a new alignment strategy using neural networks. I-TASSER was ranked as the No 1 server for protein structure prediction in recent CASP7 and CASP8 experiments. I-TASSER ONLINE - 3D models are built based on multiple-threading alignments by LOMETS and iterative TASSER simulations function inslights are then derived by matching the predicted models with protein function databases. For the modeling step, a protein 3D structure can be directly obtained from the selected template by MODELLER and displayed with global and local quality model estimation measures. Various databases such as PDB, SCOP and HOMSTRAD can be mined to find an appropriate structural template. ORION - is a web server for protein fold recognition and structure prediction using evolutionary hybrid profiles. results come by E-mail and require a viewer such as DeepView - Swiss-PdbViewer, Rasmol, or Cn3D. SWISS-MODEL - (Glaxo-Wellcome Experimental Research, Switzerland) An automated comparative protein modelling server. Databases: Subsets of the Brookhaven Protein Data Bank (PDB) database with low sequence similarity produced using the RedHom tool. RedHom: A tool to find a subset with low sequence similarity in a database. ĬPHModels (Center for Biological Sequence Analysis, Technical University of Denmark) - currently consists of the following tools: Sowhat: A neural network based method to predict contacts between C-alpha atoms from the amino acid sequence. It also incorporates a n ew ab initio folding simulation called Poing to model regions of your proteins with no detectable homology.
Phyre2 uses the alignment of hidden Markov models via HHsearch to significantly improve accuracy of alignment and detection rate. In each case I have used this site it has provide me with a model. PHYRE2 - Protein Homology/analog Y Recognition Engine - this is my favourite site for the prediction of the 3D structure of proteins. To obtain PDB coordinates for a protein of your interest, go to the Protein Data Bank or Molecules to Go or NCBI.
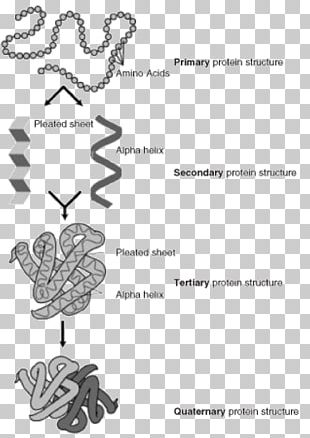
Peitsch (GlaxoWellcome, Switzerland) is here. With the two protein analysis sites the query protein is compared with existing protein structures as revealed through homology analysis.īackground: "Principles of Protein Structure, Comparative Protein Modelling and Visualization" by N. Sites are offered for calculating and displaying the 3-D structure of oligosaccharides and proteins. Online Analysis Tools - Protein Tertiary Structure PROTEIN TERTIARY STRUCTURE


 0 kommentar(er)
0 kommentar(er)
